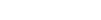A new study led by UCLA Health has identified several new risk genes for Alzheimer’s disease and progressive supranuclear palsy (PSP), a rare brain-related disorder, through a new testing approach.
Published in the journal Science, the study provides a revised model that shows how common genetic variants may collectively increase the disease risk by disrupting specific transcriptional programmes across the genome.

Discover B2B Marketing That Performs
Combine business intelligence and editorial excellence to reach engaged professionals across 36 leading media platforms.
Researchers usually depend on genome-wide association studies (GWAS), where they investigate the genomes of a large group of people to detect genetic variants that increase disease risk.
They test for markers related to a disease along the chromosome, or loci.
Commonly, each locus has dozens and sometimes hundreds or thousands of genetic markers, which are co-inherited and related to the disease.
This makes it difficult to identify the genetic markers that are actually the functional variants related to the disease.
US Tariffs are shifting - will you react or anticipate?
Don’t let policy changes catch you off guard. Stay proactive with real-time data and expert analysis.
By GlobalDataIn the new study, the authors conducted high-throughput tests to study neurodegenerative disease.
By running massively parallel reporter assays (MPRAs), they simultaneously tested 5,706 genetic variants in 25 loci that are related to Alzheimer’s and nine loci that are associated with PSP.
The authors could identify 320 functional genetic variants from the test.
They carried out a pooled CRISPR screen on 42 of those high-confidence variants in multiple types of cells to validate the results.
UCLA Gordon and Virginia MacDonald Distinguished Professor of Human Genetics, Neurology and Psychiatry and the study’s corresponding author Dr Dan Geschwind said: “We combined multiple advances that allow one to conduct high-throughput biology, in which instead of doing one experiment at a time, one does thousands of experiments in parallel in a kind of pooled format.
“This allows us approach this challenge of how to move from thousands of genetic variants associated with a disease to identifying which are functional and which genes they impact.”
The data provided evidence indicating several new Alzheimer’s risk genes, which include C4A, PVRL2 and APOC1, as well as the new risk genes PLEKHM1 and KANSL1 for PSP.
Additionally, the authors validated many previously identified risk loci and plan to study how the recently identified risk genes interact in cells and model systems.
The UCLA-Caltech MSTP training grant T32- GM008042, Rainwater Charitable Foundation grants 20180629 and A130323, National Institute of Neurological Disorders and Stroke grants 5UG3NS104095-04 and U54 NS123746, and National Institute of Aging Fellowship 1F30AG064832 provided funding for the study.